Recent genome-wide analyses in Xenopus
Recent studies utilizing genome-wide sequencing methods, chromatin immunoprecipitation and deep sequencing (ChIP-seq) and RNA deep sequencing (RNA-seq) with sequencing reads mapped to the genome, highlight the ability to interrogate genomic transcription factor binding, identify synthesized messenger RNA sequences, and observe downstream signaling responses during Xenopus development. Here is a collection of recent articles detailing these events and their importance in several key biological processes and identification of possible human disease gene associations.
____________________________
Identifying direct targets of transcription factor Rfx2 that coordinate ciliogenesis and cell movement.
Kwon et al., Genomics Data (2014), Dec 1; 2:192-194.
Click here to see the Xenbase article page.
Click here to see the article in Pubmed.
The transcription factor Rfx2 is important in ciliogenesis and cell movement of multiciliated cells. Morpholino oligos against Rfx3 were injected into X. laevis embryos at the 4-cell stage. Animal caps were taken and cultured to NF stage 20. In this article, ChIP-seq and RNA-seq analyses were performed to identify genes as direct targets of Rfx3.
____________________________
Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program.
Chiu et al., Development, (2014), Dec; 141 (23):4537-47.
Click here to see the Xenbase article page.
Click here to see the article in Pubmed.
Nodal signaling regulates mesendoderm development. RNA-seq was performed on X. tropicalis embryos with induced Foxh1 transcription factor and Nodal signaling loss of function. In this article, a combination of ChIP-seq of Foxh1 and Smad2/3 reveals an increased number of Nodal signaling target genes directly regulated by Foxh1 and Smad2/3. Foxh1 may also have Nodal signaling-independent function as well as an evolutionary conserved interaction with PouV, similar to Pou5f1-mediated regulation of human embryonic stem and epiblast cell pluripotency.
____________________________
Developmental enhancers are marked independently of zygotic Nodal signals in Xenopus.
Gupta et al., Developmental Biology (2014), Nov 1; 395 (1):38-49.
Click here to see the Xenbase article page.
Click here to see the article in Pubmed.
This article examined whether developmental enhancers were influenced by Nodal signaling during early X. tropicalis development. Active enhancer chromatin marks H3K4me1 and H3K27ac define these developmental enhancers. These marks are seen as early as blastula stage and that Smad2/3 only strongly associates with these regions at gastrula stages. These developmental enhancer marks remain even after embryonic perturbation of Nodal signaling using the drug SB431542. These enhancers seem to be in an active conformation prior to and are established before Nodal signaling. The authors suggest that many developmental enhancers are prepared maternally before extrinsic signaling exposure occurs.
____________________________
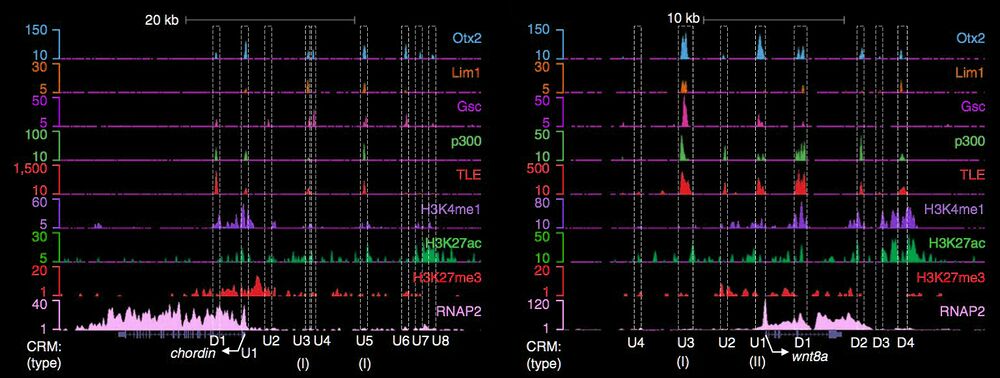
Occupancy of tissue-specific cis-regulatory modules by Otx2 and TLE/Groucho for embryonic head specification.
Yasuoka et al., Nature Communications (2014) Jul 9; 5:4322.
Click here to see the Xenbase article page.
Click here to see the article in Pubmed.
Head specification by the transcription factor Orthodenticle (otx) is highly conserved among bilateria. ChIP-seq and RNA-seq in X. tropicalis gastrulae were performed to identify direct Otx2 and TLE/Groucho mediated cis-regulatory modules (CRMs). This article describes two distinct types of Otx2- and TLE-occupied CRMs that can be identified marking target genes regulated during head specification by the Xenopus head organizer.
____________________________
Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis.
Wills et al., Methods (2014), Apr 1; 66 (3): 410-21.
Click here to see the Xenbase article page.
Click here to see the article in Pubmed.
Chromatin immunoprecipitation and deep seqhencing (ChIP-seq) is a powerful tool for identifying chromatin remodeling factors, histone modifications, and direct genomic targets for transcription factors. This article provides a detailed ChIP-seq analysis protocol for use in X. tropicalis and X. laevis embryology and cell biology.
