Xenopus animal cap transcriptome
Nucleic Acids Research, Volume 46, Issue 17, 28 September 2018, Pages 8772–8787, https://doi.org/10.1093/nar/gky771.
Building a mucociliary epithelium
[click image to enlarge]
Angerilli et al. investigated the dynamics of the transcriptional landscape in X. tropicalis Animal Cap explants during their differentiation into mucociliary epithelium. Providing an online Web Tool, they detail global changes in mRNA expression, an unanticipated diversity of mRNA splicing isoforms, expression of repetitive DNA elements and the complexity of circular RNAs. Computational analysis of transcription factor hubs predicts novel genetic drivers for epidermal differentiation. Their findings are published in Nucleic Acids Research.
Click the link to the Web Tool:
Click the link to the expression profiling by high throughput sequencing on GEO:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111171
Click here to view article at Nucleic Acids Research.
Click here to view article on Pubmed.
Click here to view article on Xenbase.
ABSTRACT
With the availability of deep RNA sequencing, model organisms such as Xenopus offer an outstanding opportunity to investigate the genetic basis of vertebrate organ formation from its embryonic beginnings. Here we investigate dynamics of the RNA landscape during formation of the Xenopus tropicalis larval epidermis. Differentiation of non-neural ectoderm starts at gastrulation and takes about one day to produce a functional mucociliary epithelium, highly related to the one in human airways. To obtain RNA expression data, uncontaminated by non- epidermal tissues of the embryo, we use prospective ectodermal explants called Animal Caps (ACs), which differentiate autonomously into a ciliated epidermis. Their global transcriptome is investigated at three key timepoints, with a cumulative sequencing depth of ∼108 reads per developmental stage. This database is provided as online Web Tool to the scientific community. In this paper, we report on global changes in gene expression, an unanticipated diversity of mRNA splicing isoforms, expression patterns of repetitive DNA Elements, and the complexity of circular RNAs during this process. Computationally we derive transcription factor hubs from this data set, which may help in the future to define novel genetic drivers of epidermal differentiation in vertebrates.
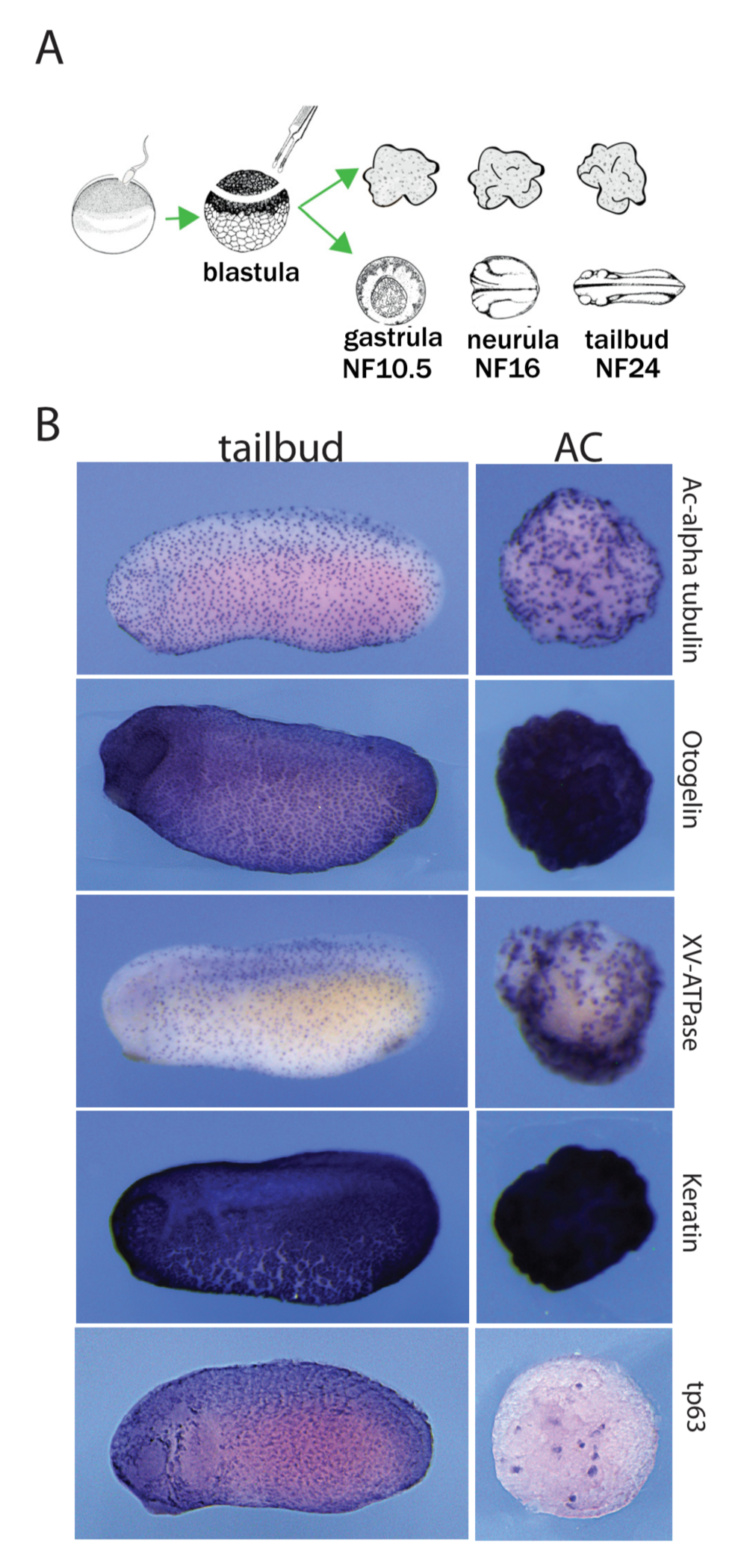
Figure 1. Autonomous differentiation of AC explants into a mucocil- iary epithelium. (A) Experimental procedure and timeline. (B) Early tail- bud embryos and sibling ACs are stained for typical epidermal markers: acetylated-alpha tubulin protein (ICC), and for otogelin, xv-atpase, ker- atin and tp63 mRNA (RNA in situ hybridisation). ACs show a staining pattern comparable to the embryonic skin.
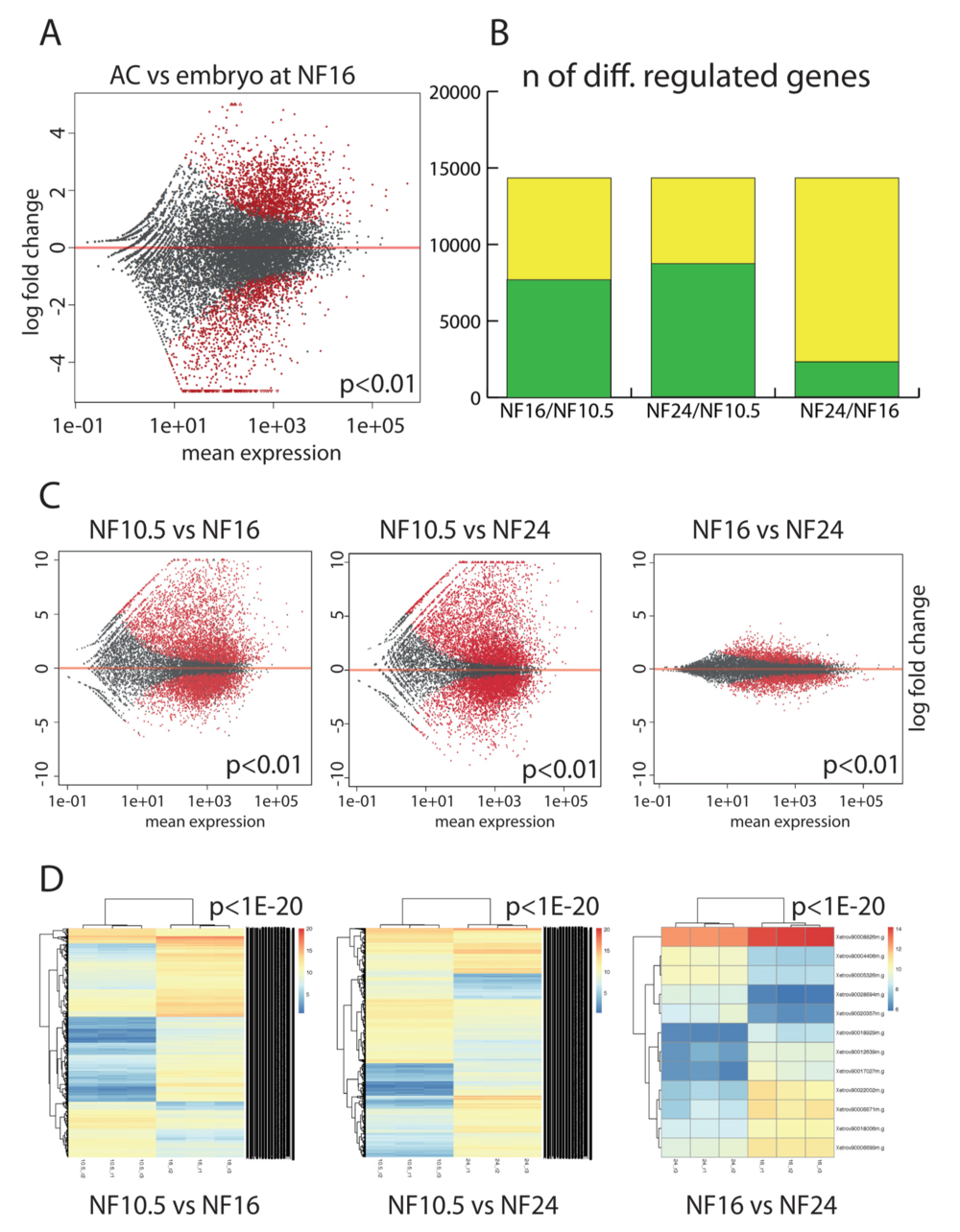
Figure 2. Comparativetranscriptomeanalysisofepidermaldifferentiation.(A)GeneexpressionintheACversuswholeembryoattheneurulastage(NF16). Each dot represents a gene. In red, genes that are differentially expressed in a significant manner (P < 0.01), in grey genes with not-significant variation. (B) Plotting the extent of differential gene expression in AC. Yellow: total number of genes detected in the RNAseq. Green: genes that are differentially regulated between the given developmental stages (P < 0.01). The maximal number is defined by the annotated gene models present in XenTro9.1. (C) Scatter plots detailing the extent and magnitude of differential gene expression between indicated developmental stages (P < 0.01). (D) Unsupervised clustering of the differentially expressed genes from the biological replicates with a cutoff of P < 1e–20.
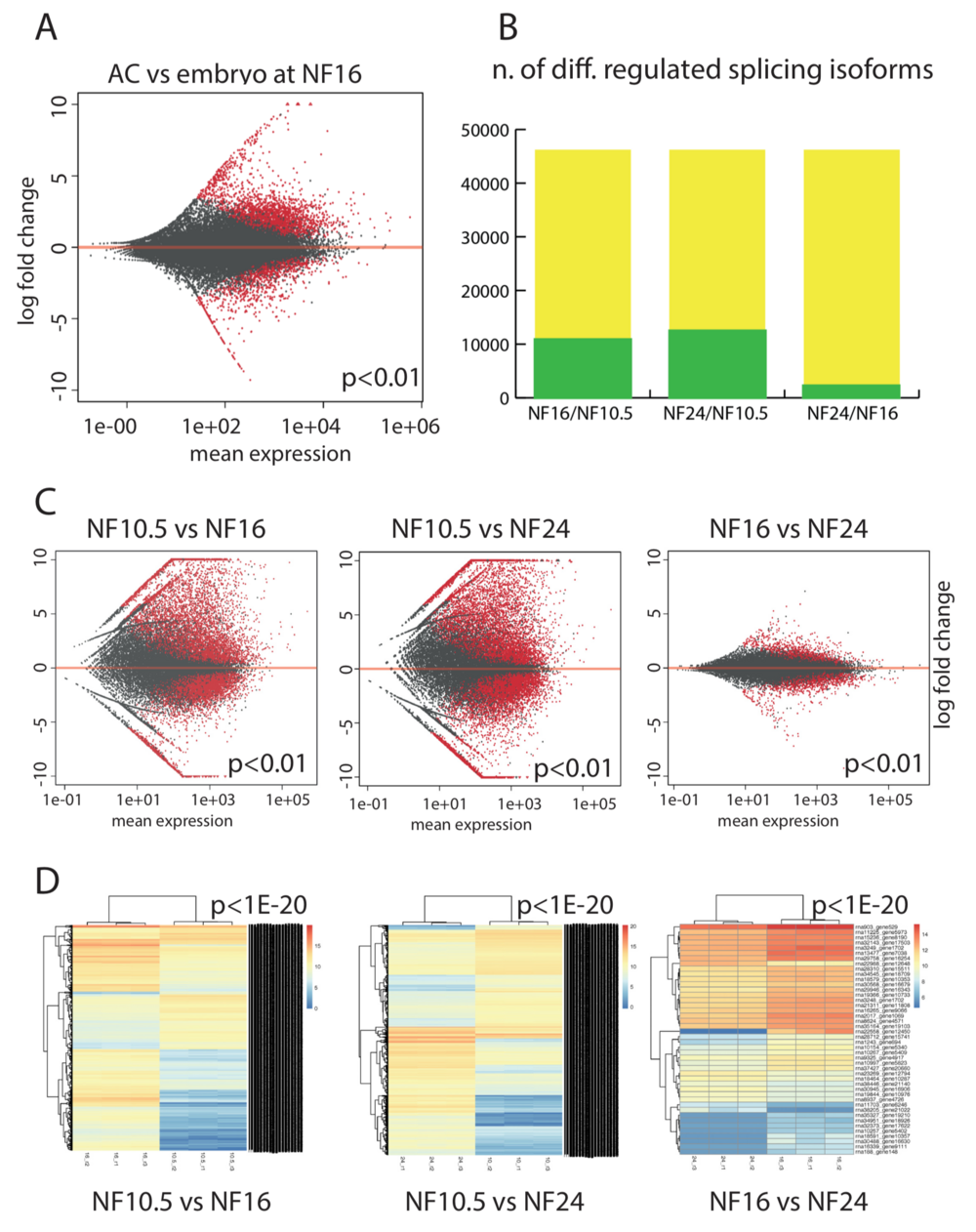
Figure 3. Epidermis-specific splicing isoforms. (A) Expression of splicing isoforms in the AC vs whole embryo at the neurula stage (NF16). Each dot represents an annotated splicing isoform. In red, splice isoforms that are differentially expressed in a significant manner (P < 0.01), in grey transcripts with not-significant variation. (B) Plotting the extent of differentially expressed splice isoforms in AC. Green: isoforms that are differentially regulated between the given developmental stages (P < 0.01). Yellow: the maximal number is defined by the annotated transcript models present in NCBI Xenopus tropicalis annotation release 103. (C) Scatter plots detailing the extent and magnitude of differential splice isoform expression between indicated developmental stages (P < 0.01). D) Unsupervised clustering of the differentially expressed splicing isoforms from the biological replicates with a cutoff of P < 1e–20.
Adapted with permission from Oxford University Press on behalf of Nucleic Acids Research: Angerilli et al. (2018). The Xenopus animal cap transcriptome: building a mucociliary epithelium. Nucleic Acids Research, Volume 46, Issue 17, 28 September 2018, Pages 8772–8787, https://doi.org/10.1093/nar/gky771. Copyright (2018).
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2018-10-08