A systemic cell cycle block impacts stage-specific histone modification profiles during Xenopus embryogenesis
Daniil Pokrovsky, Ignasi Forné, Tobias Straub, Axel Imhof, Ralph A W Rupp
PLoS Biol. 2021 Sep 7;19(9):e3001377. doi: 10.1371/journal.pbio.3001377. eCollection 2021 Sep.
Click here to view article at PLoS Biology.
Click here to view article at Pubmed.
Click here to view article on Xenbase.
Abstract
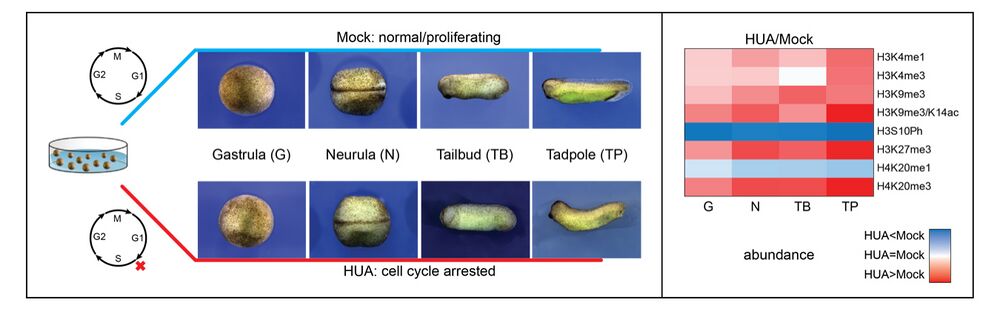
Forming an embryo from a zygote poses an apparent conflict for epigenetic regulation. On the one hand, the de novo induction of cell fate identities requires the establishment and subsequent maintenance of epigenetic information to harness developmental gene expression. On the other hand, the embryo depends on cell proliferation, and every round of DNA replication dilutes preexisting histone modifications by incorporation of new unmodified histones into chromatin. Here, we investigated the possible relationship between the propagation of epigenetic information and the developmental cell proliferation during Xenopus embryogenesis. We systemically inhibited cell proliferation during the G1/S transition in gastrula embryos and followed their development until the tadpole stage. Comparing wild-type and cell cycle-arrested embryos, we show that the inhibition of cell proliferation is principally compatible with embryo survival and cellular differentiation. In parallel, we quantified by mass spectrometry the abundance of a large set of histone modification states, which reflects the developmental maturation of the embryonic epigenome. The arrested embryos developed abnormal stage-specific histone modification profiles (HMPs), in which transcriptionally repressive histone marks were overrepresented. Embryos released from the cell cycle block during neurulation reverted toward normality on morphological, molecular, and epigenetic levels. These results suggest that the cell cycle block by HUA alters stage-specific HMPs. We propose that this influence is strong enough to control developmental decisions, specifically in cell populations that switch between resting and proliferating states such as stem cells.
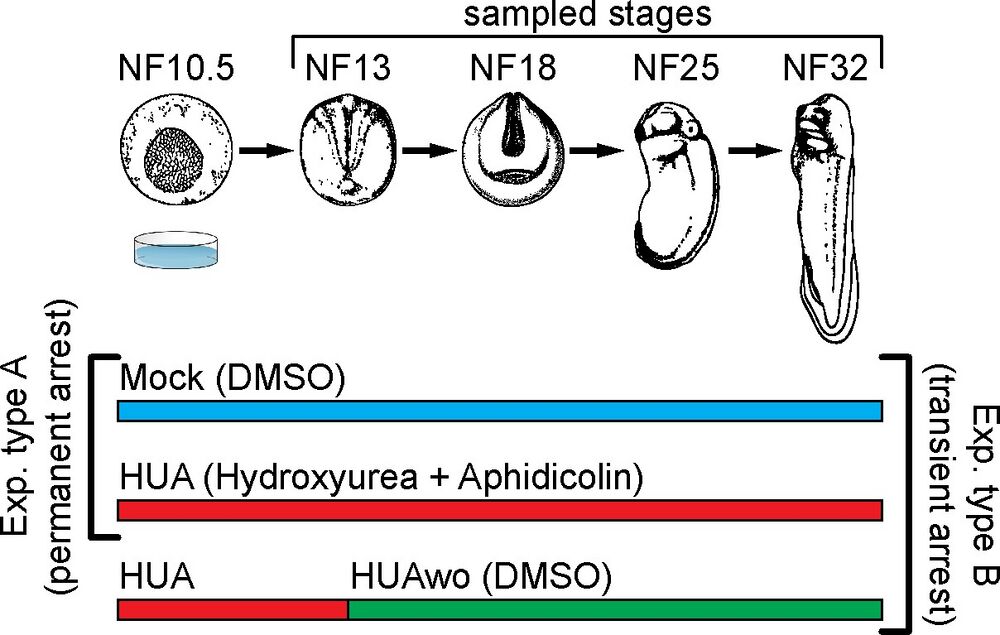
Fig 1. Experimental design. Top part: a schematic representation of Xenopus laevis embryonic development. Developmental stages (NF) according to Nieuwkoop and Faber (1994). Stages used for mass spectrometry of histone modifications and embryonic analyses are characterized by the following features: NF13—late gastrula, germ layer specified; NF18—neurula, germ layer patterning and differentiation; NF25—tailbud stage, organogenesis; NF32—early tadpole, body plan established. Bottom part: We perform 2 types of experiments: Experiment type A (G1/S block)—embryos are split in 2 groups, which from NF10.5 on are continuously incubated in HUA solution or control solution (DMSO as carrier). Experiment type B (transient arrest)—embryos are split in 3 groups: continuous Mock, continuous HUA, and transient HUA. In the last group, HUA solution is replaced at NF13 with DMSO solution. DMSO, dimethyl sulfoxide; HUA, hydroxyurea and aphidicolin; HUAwo, HUA washout. https://doi.org/10.1371/journal.pbio.3001377.g001
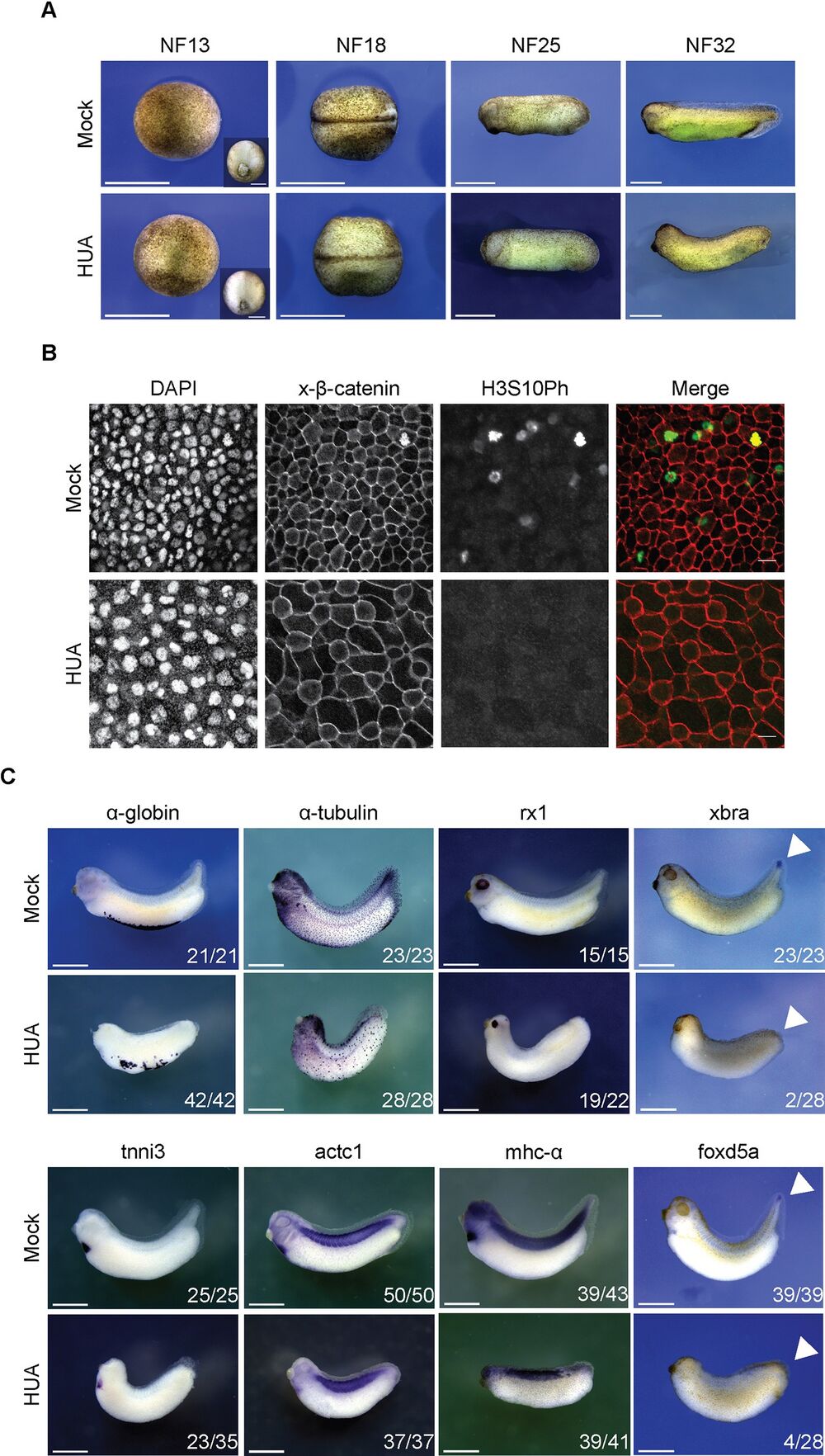
Fig 3. Morphological development of HUA-arrested embryos. (A) During gastrulation, Mock and HUA embryos are indistinguishable. At NF18, the latter show a delay in neural tube closure. More severe malformations are detectable at stages NF25 and NF32, most notably reduced eye formation and absence of tail bud. Scale bars: 1 mm. (B) Cell size at the tailbud stage. Flattened Z-stack images show fields from embryonic skin at constant magnification (scale bars: 20 μm). Immunofluorescence detects cell borders (beta-catenin), nuclei (DAPI), and mitotic cells (H3S10Ph). GIFs from Z-stacks are presented in the Supporting information (S1 and S2 Videos). (C) Whole mount RNA in situ hybridization for indicated marker genes. Images are representative for the majority of Mock- or HUA-treated embryos from 3 biological experiments. While mRNAs of α-tubulin and rx1 (skin, neuronal), α-globin, tnni3, actc1, and mhc-α (mesodermal) are detected in both conditions, xbra and foxd5a (tail blastema) are absent in HUA embryos. Numbers indicate embryos positive for the marker over the total number of analyzed embryos. Scale bars: 1 mm. HUA, hydroxyurea and aphidicolin. https://doi.org/10.1371/journal.pbio.3001377.g003
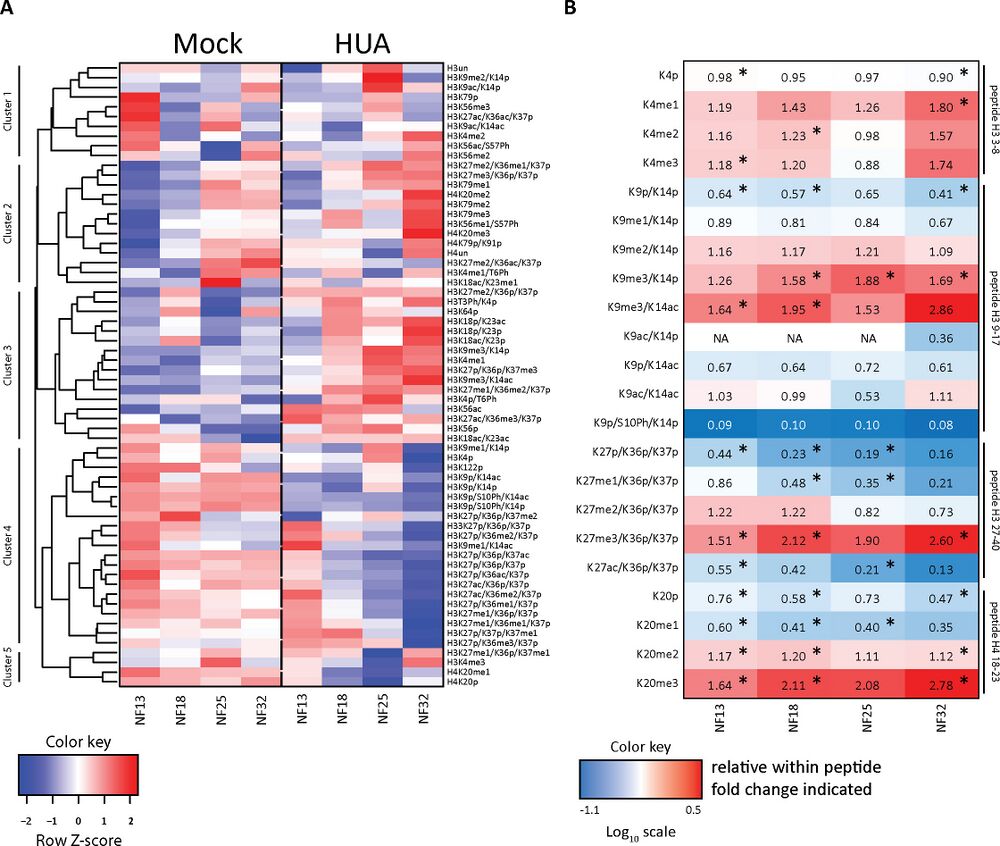
Fig 4. Mitotic activity shapes stage-specific HMPs. (A) Heatmap of the absolute histone modification states abundance, normalized to the corresponding R10 spiketides. Data in columns represent an average from 3 biological replicates/condition. Color key: row Z-score. The individual quantitative observations can be found in S2 Data. (B) Ratio heatmap of relative histone modifications abundance for selected histone marks. In the first step, relative distributions of the indicated modification states were calculated in percentages within each tryptic peptide (see S4 Fig). Then, HUA values were divided by Mock values to produce the relative change for each histone modification state between the 2 conditions. Color key is based on the Log10 scale. Numerical values in the cells indicate the fold-change. Values greater than 1.0 indicate an increase in HUA condition; values smaller than 1.0 indicate a higher abundance in Mock condition. NA, not applicable, when values in Mock were 0. Asterisks (*), adj.p < 0.1 (Benjamini–Hochberg procedure). “p,” propionylated (naturally unmodified); “un,” tryptic peptide, which has no modification state. The direct comparison of relative histone PTMs abundance is shown in S4 Fig, with p-values indicated in S5 Table. HMP, histone modification profile; HUA, hydroxyurea and aphidicolin; PTM, posttranslational modification of histone. https://doi.org/10.1371/journal.pbio.3001377.g004
Adapted with permission from PLoS on behalf of PLoS Biology: Pokrovsky, et al. (2021). A systemic cell cycle block impacts stage-specific histone modification profiles during Xenopus embryogenesis. PLoS Biol. 2021 Sep 7;19(9):e3001377. doi: 10.1371/journal.pbio.3001377. eCollection 2021 Sep.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2021-11-09