Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex
Xuechen Zhu, Gaoxingyu Huang, Chao Zeng, Xiechao Zhan, Ke Liang, Qikui Xu, Yanyu Zhao, Pan Wang, Qifan Wang, Qiang Zhou, Qinghua Tao, Minhao Liu, Jianlin Lei, Chuangye Yan, Yigong Shi
Science. 2022 Jun 10;376(6598):eabl8280. doi: 10.1126/science.abl8280. Epub 2022 Jun 10.
Click here to view article at Science.
Click here to view article at PubMed.
Click here to view article on Xenbase.
Abstract
The nuclear pore complex (NPC) mediates nucleocytoplasmic cargo transport. Here, we present a single-particle cryo–electron microscopy reconstruction of the cytoplasmic ring (CR) subunit from the Xenopus laevis NPC at 3.7- to 4.7-angstrom resolution. The structure of an amino-terminal domain of Nup358 has been resolved at 3.0 angstroms, facilitating the identification of five Nup358 molecules in each CR subunit. Our final model of the CR subunit included five Nup358, two Nup205, and two Nup93 molecules in addition to the two previously characterized Y complexes. The carboxyl-terminal fragment of Nup160 served as an organizing center at the vertex of each Y complex. Structural analysis revealed how Nup93, Nup205, and Nup358 facilitate and strengthen the assembly of the CR scaffold that is primarily formed by two layers of Y complexes.
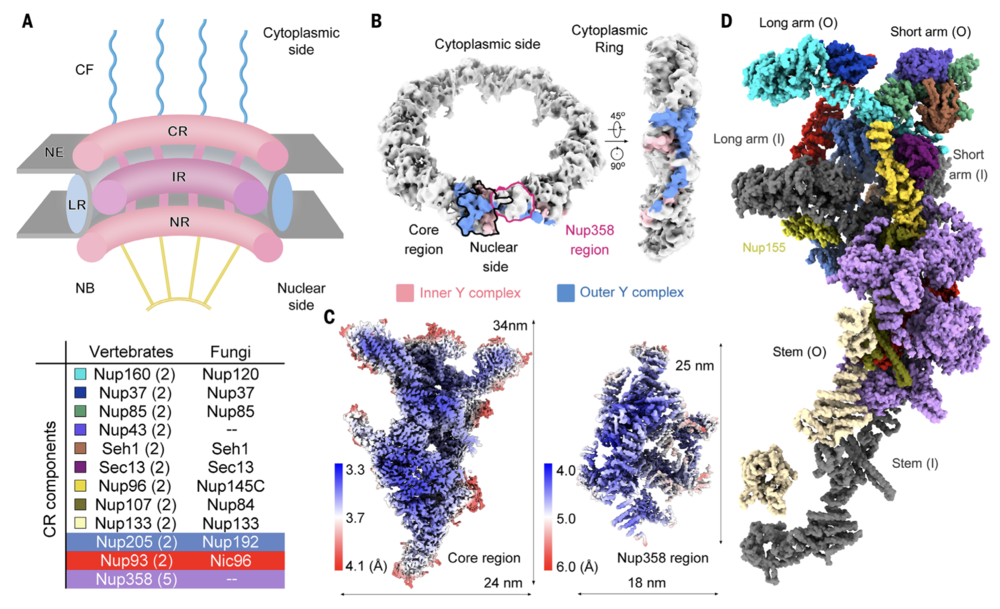
Fig. 1. Single-particle cryo-EM analysis of the CR subunit of the X. laevis NPC. (A) Schematic overview of the architecture of a vertebrate NPC. Shown is a cut-open side view of a cartoon depicting an NPC embedded in the NE. Each NPC, along its cytonuclear axis, comprises the CF, CR, IR, NR and NB. A LR localized inside the lumen of the NE encompasses the IR. The CR components from vertebrates and fungi are listed in the table below. The copy number of each CR component in a vertebrate NPC is indicated in brackets. (B) EM map of the CR at 22.2-Å resolution. The core region and the Nup358 region in one CR subunit, which are separately calculated as single particles, are indicated by black and magenta contours, respectively, in the left panel. The densities corresponding to the inner and outer Y complexes in one CR subunit are colored pink and blue, respectively. (C) Heatmaps for the local resolutions of the core region and the Nup358 region that were separately reconstructed to average resolutions of 3.7 and 4.7 Å, respectively. The local resolutions were calculated in RELION3.0 (59) and presented in ChimeraX (58). (D) Overall structure of a CR subunit from the X. laevis NPC. The structure is presented in approximately the same view as the two Y complexes in the right panel of (B). Protein components of the outer Y complex and Nup205, Nup93, and Nup358 molecules are colored the same as those annotated in (A). The inner Y complex is colored dark gray. Nup155, which connects the CR to the IR, is labeled but not included in the table in (A).
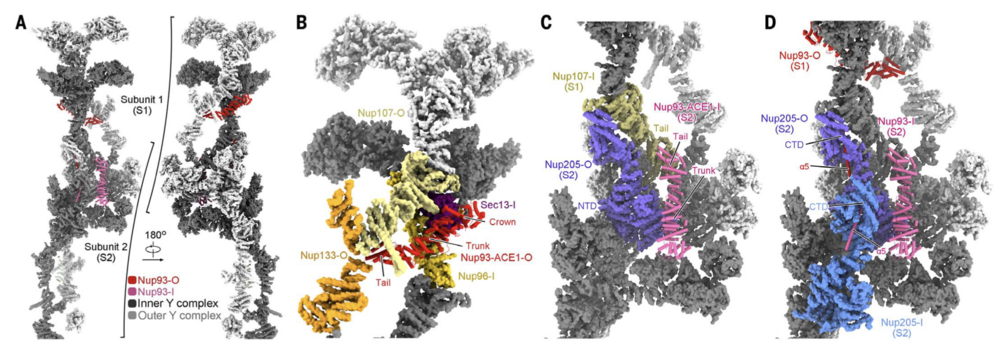
Fig. 4. Nup93 bridges Y complexes and Nup205 molecules. (A) Nup93 molecules bridge adjacent Y complexes as well as Nup205 molecules. Identical components from two adjacent CR subunits, designated S1 and S2, are presented. Nup205 and the Y complexes are shown in surface representation. Inner and outer components are colored dark and light gray, respectively. Two copies of Nup93 are present in each CR subunit, with the one closer to the central pore designated as Nup93-I (hot pink) and the other as Nup93-O (red). For visual clarity, only Nup93-O from S1 and Nup93-I from S2 are shown. (B) Nup93-ACE1-O connect the stems of the inner and outer Y complexes in each CR subunit. The bridging role is achieved through direct binding to the ACE1 domains in Nup96-I and Nup107-O. These proteins form a triangular ACE1 core at the stem of the Y complexes, which is bolstered by Sec13-I and Nup133-O. Previously defined module names of the ACE1 domain, crown, trunk, and tail are labeled. (C) Nup93- ACE1-I bridges two adjacent subunits through direct binding to the ACE1 of Nup107-I from S1 and the NTD of Nup205-O from S2. (D) The a5 helix from Nup93 inserts into the axial groove of Nup205-CTD. The structures, shown as cartoon or surface, were prepared in ChimeraX.
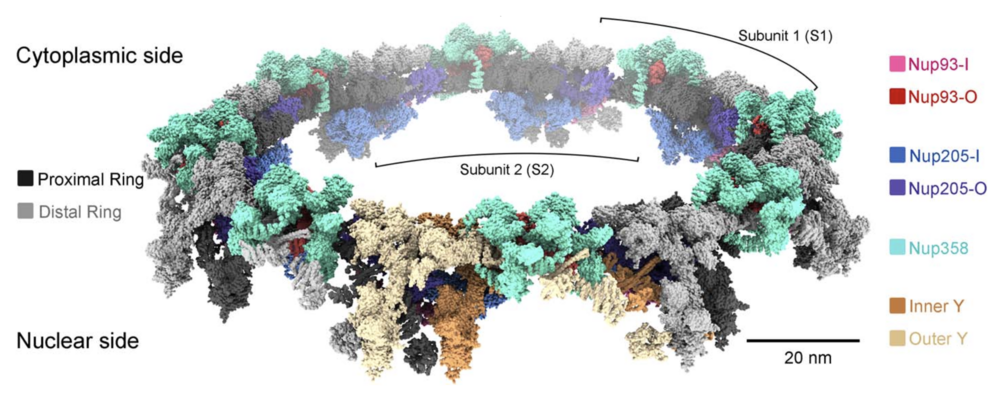
Fig. 6. Structure of the double-layered CR scaffold. Shown is a composite model of the X. laevis CR. The model is shown as surface in ChimeraX. In addition to the two concentric Y-complex rings that each comprises eight head-to-tail assembled Y complexes, our present study identifies Nup358, Nup205, and Nup93 to be constituents of the CR scaffold. The inner and outer Y complexes in the subunit closest to reader are colored sandy brown and wheat, respectively, and in the other seven subunits they are colored dark and light gray, respectively.
Adapted with permission from American Association for the Advancement of Science on behalf of Science: Zhu et al. (2022). Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex. Science. 2022 Jun 10;376(6598):eabl8280. doi: 10.1126/science.abl8280. Epub 2022 Jun 10.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2022-07-13