Critical histone modifier for multiciliated cell differentiation identified
The histone H4K20 methyltransferase SUV4-20H1/KMT5B is required for multiciliated cell differentiation in Xenopus
Angerilli A, Tait J, Berges J, Shcherbakova I, Pokrovsky D, Schauer T, Smialowski P, Hsam O, Mentele E, Nicetto D, Rupp RA
Life Sci Alliance 2023 Jul 01;67:. doi: 10.26508/lsa.202302023.
Click here to view article at Life Science Alliance.
Click here to view article on PubMed.
Click here to view article on Xenbase.

Significance
The Xenopus larval skin is a prime model for the mucociliary epithelium found in human airways. Hallmarks of these epithelia are multiciliated cells (MCCs), whose coordinated cilia strokes generate directional fluid flow. Recent work, mostly in Xenopus, has established that cilia tufts are formed by a coordinated transcriptional upregulation of hundreds of genes in postmitotic MCC precursor cells. This transcriptional burst is needed to produce the building blocks of the cilia tuft – centriole-related basal bodies, a subcortical Actin meshwork, and motile axonemes, emanating from membrane-anchored basal bodies.
The publication by Angerilli, Tait and colleagues from the Rupp lab now indicates that normal ciliogenesis requires a permissive chromatin landscape consisting primarily of nucleosomes decorated with dimethylated Histone H4 Lysine 20 (H4K20me2). This modification is deposited on chromatin by the related histone methyltransferases SUV4-20H1/KMT5B and SUV4-20H2/KMT5C, which use H4K20me1 as substrate. Genome-wide transcriptome profiling of ectodermal explants shows that transcription of hundreds of ciliogenic genes is significantly downregulated upon SUV4-20H1 depletion. This results in severe structural defects in the cell, as well as compromised fluid flow at the embryonic surface. Cilia can be restored by overexpressing catalytically active (but not inactive) SUV4-20H1 protein, or by overexpression of PHF8/KDM7B, a histone demethylase known to preferentially remove the methyl group from H4K20me1 at gene promoters. Surprisingly, knocking down SUV4-20H2 has no effect on MCC differentiation, although this enzyme contributes to H4K20 di-methylation and is needed to generate H4K20 tri-methylation. Taken together, this indicates that the conversion of H4K20me1 to H4K20me2 by SUV4-20H1/KMT5B is critical for the formation of cilia tufts.
Abstract
H4 lysine 20 dimethylation (H4K20me2) is the most abundant histone modification in vertebrate chromatin. It arises from sequential methylation of unmodified histone H4 proteins by the mono-methylating enzyme PR-SET7/KMT5A, followed by conversion to the dimethylated state by SUV4-20H (KMT5B/C) enzymes. We have blocked the deposition of this mark by depleting Xenopus embryos of SUV4-20H1/H2 methyltransferases. In the larval epidermis, this results in a severe loss of cilia in multiciliated cells (MCC), a key component of mucociliary epithelia. MCC precursor cells are correctly specified, amplify centrioles, but ultimately fail in ciliogenesis because of the perturbation of cytoplasmic processes. Genome-wide transcriptome profiling reveals that SUV4-20H1/H2-depleted ectodermal explants preferentially down-regulate the expression of several hundred ciliogenic genes. Further analysis demonstrated that knockdown of SUV4-20H1 alone is sufficient to generate the MCC phenotype and that its catalytic activity is needed for axoneme formation. Overexpression of the H4K20me1-specific histone demethylase PHF8/KDM7B also rescues the ciliogenic defect in a significant manner. Taken together, this indicates that the conversion of H4K20me1 to H4K20me2 by SUV4-20H1 is critical for the formation of cilia tufts.

Figure 1. SUV4-20H1/H2 enzymes are required for ciliogenesis. (A, A’) Relative abundance of H4K20 methyl states by mass spectrometry in X. laevis bulk embryonic chromatin upon control (CoMo), single or double KD of SUV4-20H1 and SUV4-20H2 enzymes. (B, C) Representative immunocytochemistry images of multiciliated cells (acetylated a-tubulin) in a tail bud stage embryo upon control or SUV4-20H dKD (inserts show enlarged sections of the same images, scale bars = 1 mm) and (C) quantification of (B) (n = 6 biological replicates). (D, E) Representative immunofluorescence images detailing the multiciliated cell phenotype upon suv4-20h1/2 KD. Basal bodies are green (hyls1-GFP), the cilia are magenta (acetylated a-tubulin antibody), the apical actin meshwork is red (phalloidin) and the nucleus is blue (DAPI). (E) Confocal analysis of the actin meshwork and docked basal bodies in CoMo or H1H2Mo-injected MCC. Panels show: 1—a representative MCC, 2—overlap between the actin cap and basal bodies on the apical surface, 3—basal bodies in only the uppermost Z-sections, 4—an apical view of the basal bodies, 5—a deep Z-section close to the cell nucleus. (F, G) Quantification of the number of MCCs showing reduced cilia and filamentous actin staining after confocal analysis. We measured 144 CoMo and 163 H1H2Mo-injected multiciliated cells. Error bars represent standard deviations.
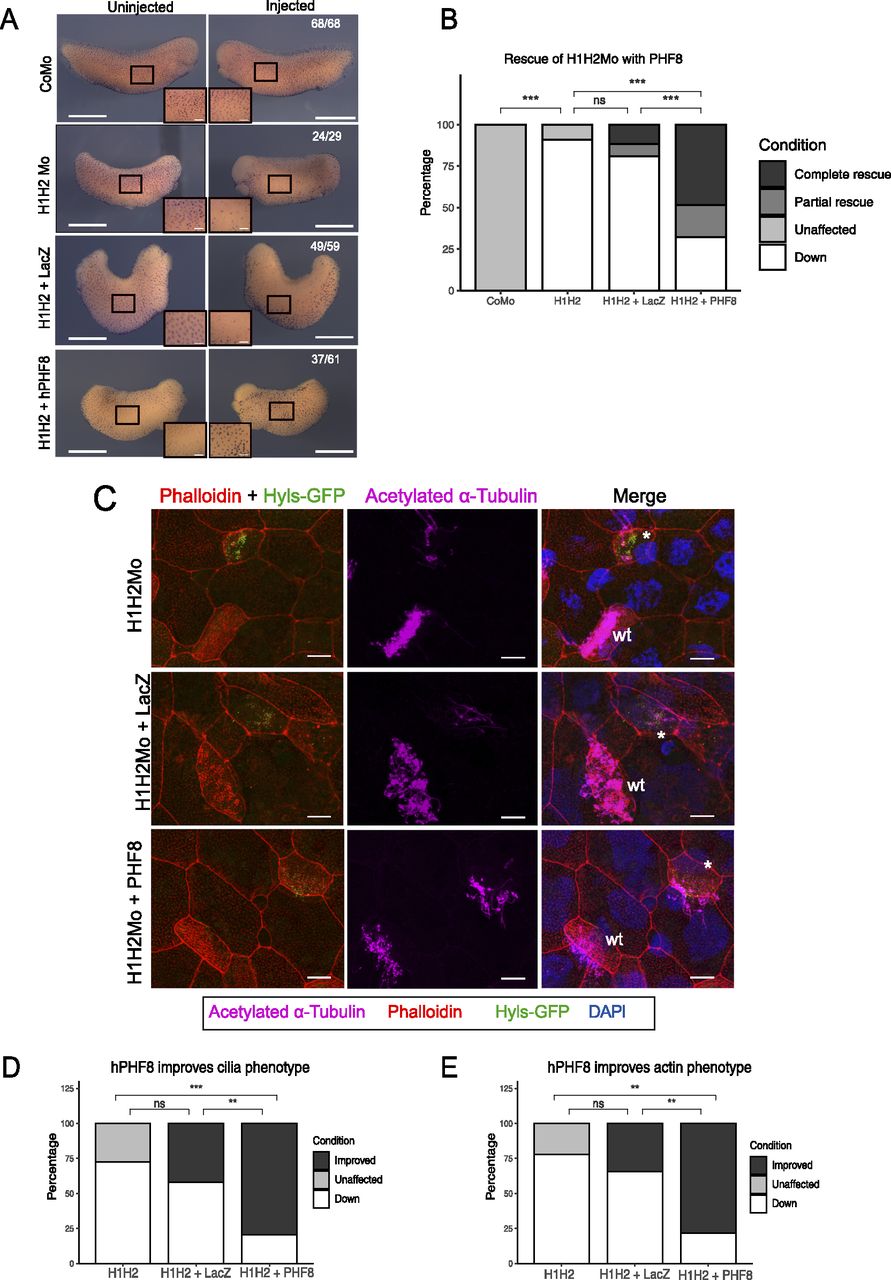
Figure 3. Rescue of the ciliogenic phenotype with hPHF8. (A) Representative immunocytochemistry images of tail bud stage embryos stained for acetylated a-tubulin (multiciliated cells). Injected reagents shown on y-axis, uninjected, and injected sides shown on top. Scale bars = 1 mm (whole embryo) and 200 μm (inserts), n = 4 biological replicates. (B) Quantification of (A). (C) Representative confocal images detailing the multiciliated cell phenotype upon suv4-20h1/2 KD and both rescue conditions. The basal bodies are green (hyls1-GFP), the ciliary axonemes are magenta (acetylated a-tubulin antibody), the apical actin meshwork is red (phalloidin), and the nucleus is blue (DAPI). A mosaic injection scheme is used allowing KD and wt cells to be present in the same field of view (* = KD MCCs, wt = wildtype MCCs). (C, D, E) Quantification of (C). Scale bars = 10 μm, n = 3 biological replicates.
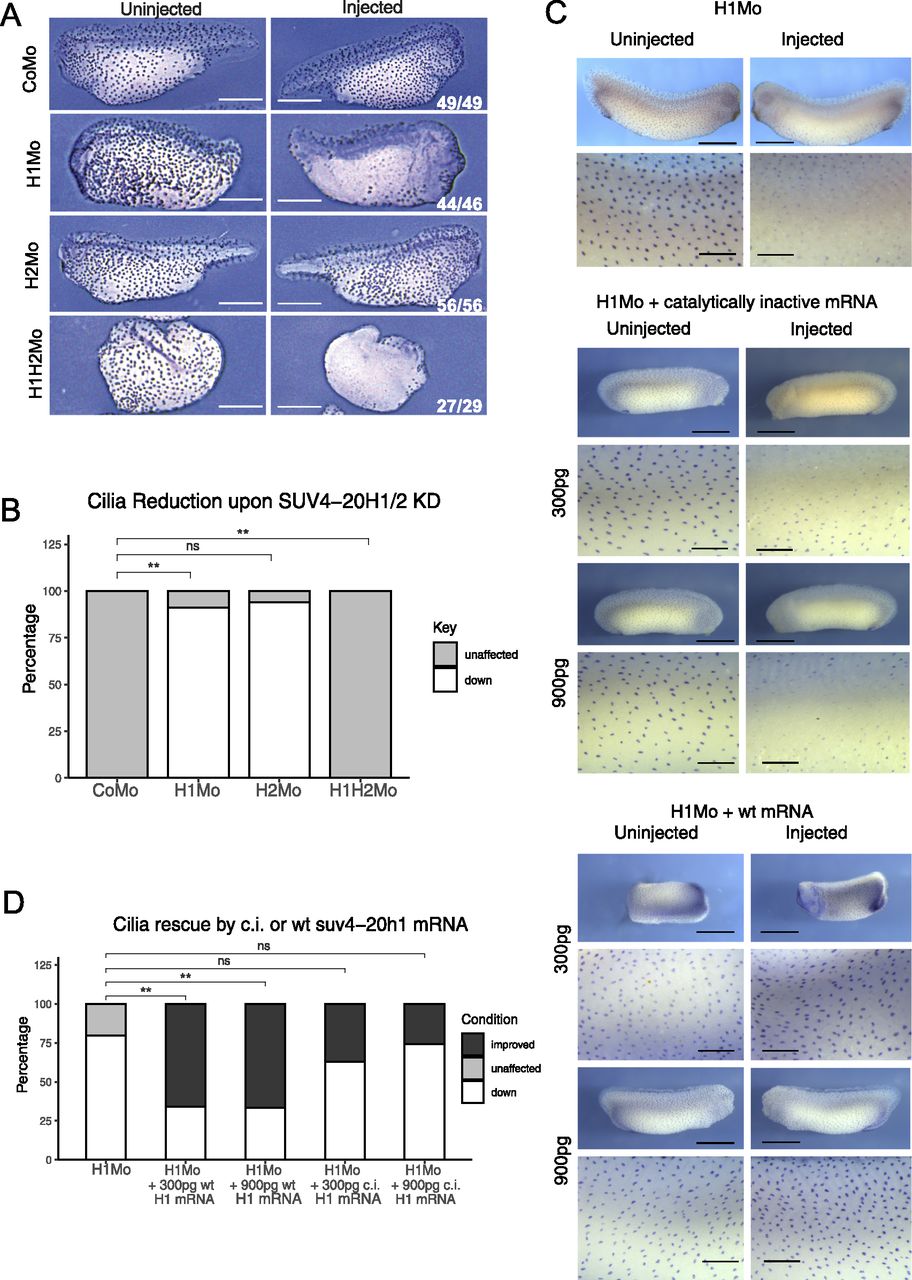
Figure 5. SUV4-20H1 activity is needed for cilia tuft formation. Representative immunostainings of multiciliated cells (acetylated a-tubulin) on the tail bud stage embryos. (A) X. tropicalis embryos upon KD of the suv4-20h enzymes individually or in concert. Scale bars = 1 mm, n = 3 biol. (A, B) replicates (B) Quantification of the experiment in (A). (C) X. laevis embryos upon KD of suv4-20h1 and rescued with either catalytically inactive (c.i.) or WT suv4-20h1 mRNA. Scale bars = 1 mm (whole embryo) and 200 μm (inserts), n = 3 biological replicates. (C, D) Quantification of panel (C).
Adapted with permission from Life Science Alliance on behalf of Life Science Alliances: Angerilli & Tait et al. (2023). The histone H4K20 methyltransferase SUV4-20H1/KMT5B is required for multiciliated cell differentiation in Xenopus. Life Sci Alliance 2023 Jul 01;67:. doi: 10.26508/lsa.202302023.
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
Last Updated: 2023-08-08